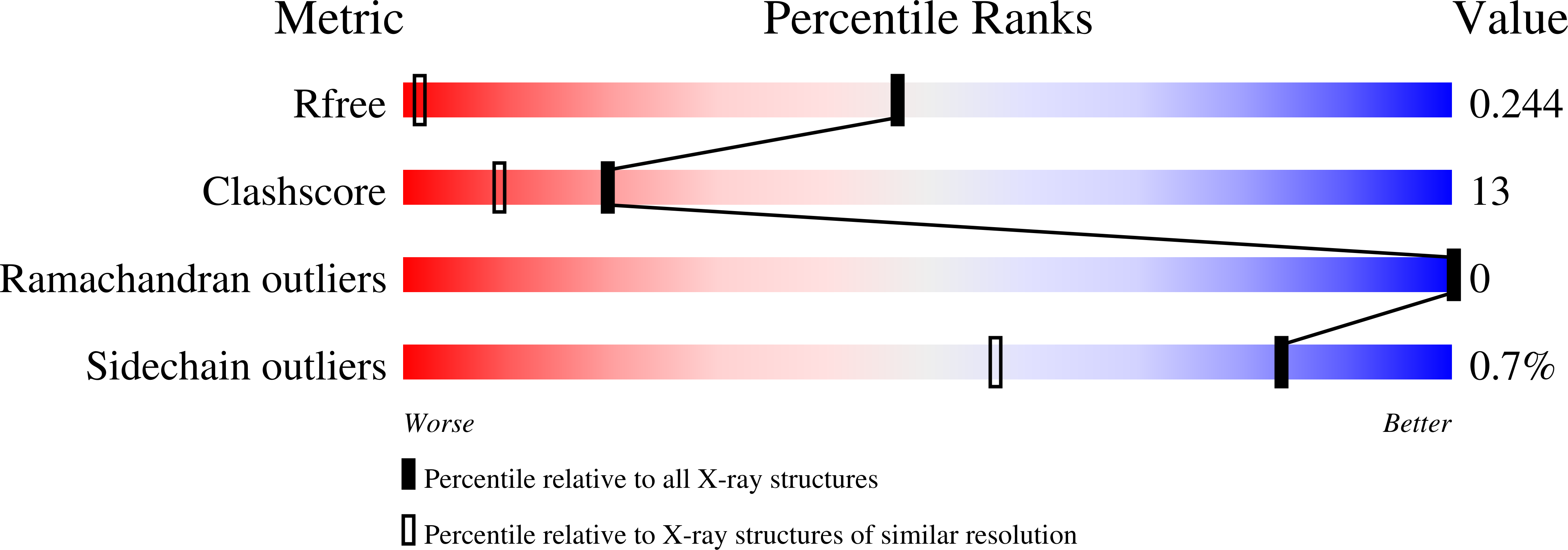
Regulation of Neurexin 1beta Tertiary Structure and Ligand Binding through Alternative Splicing
Shen, K.C., Kuczynska, D.A., Wu, I.J., Murray, B.H., Sheckler, L.R., Rudenko, G.(2008) Structure 16: 422-431
- PubMed: 18334217
- DOI: https://doi.org/10.1016/j.str.2008.01.005
- Primary Citation of Related Structures:
2R16, 2R1B, 2R1D - PubMed Abstract:
Neurexins and neuroligins play an essential role in synapse function, and their alterations are linked to autistic spectrum disorder. Interactions between neurexins and neuroligins regulate inhibitory and excitatory synaptogenesis in vitro through a "splice-insert signaling code." In particular, neurexin 1beta carrying an alternative splice insert at site SS#4 interacts with neuroligin 2 (found predominantly at inhibitory synapses) but much less so with other neuroligins (those carrying an insert at site B and prevalent at excitatory synapses). The structure of neurexin 1beta+SS#4 reveals dramatic rearrangements to the "hypervariable surface," the binding site for neuroligins. The splice insert protrudes as a long helix into space, triggers conversion of loop beta10-beta11 into a helix rearranging the binding site for neuroligins, and rearranges the Ca(2+)-binding site required for ligand binding, increasing its affinity. Our structures reveal the mechanism by which neurexin 1beta isoforms acquire neuroligin splice isoform selectivity.
Organizational Affiliation:
Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109, USA.